
The terrible COVID pandemic brought attention to zoonotic spillovers. The main idea is that SARS-CoV-2, the virus that causes COVID-19, changed in bats and then spread to other wild animals sold alive for food in the Wuhan markets, where they ultimately infected humans. The rest is, as they say, history.
We've had to deal with many zoonotic diseases before, like Ebola, avian and swine influenzas. But it’s easy to forget that microbes do not only spread from animals to humans. It’s a two-way street — and this can be surprising.
Researchers at the University College London (UCL) have now studied viral dynamics in great detail and discovered: people give twice as many viruses to domesticated and wild animals as we get from them.
“Anthroponotics (human-to-animal diseases) are far more common than we thought with humans acting more as a source than a sink in the flow of viral exchange between host species,” Professor Francois Balloux of UCL and co-author of the new study told ZME Science.
The two-way street of viral infection
Anthroponosis, or the jump of viruses from humans to other animals, is increasingly being detected. Examples of such “reverse zoonotic” events include epidemics of the 2009 influenza A (H1N1) pandemic virus in pigs and epidemics of SARS-CoV-2 in minks and white-tailed deer. Pet owners may be more aware of this dynamic as it is common for dogs, cats, and even ferrets to show flu-like symptoms after close contact with infected humans.
The importance of humans as a viral source was never clearly understood. And, of course, there’s a bias in this kind of research to focus on animal-to-human transmission. For their study, the researchers set about the herculean task of analyzing nearly 12 million viral genomes. These were used to build phylogenetic trees to help them find patterns of previous host jumps.
Imagine a scenario where a virus is found in both bats and humans, and the viral strains from these two hosts differ by only a few mutations. This similarity suggests a recent transmission event. But the critical question is: who infected whom? That’s what the phylogenetic trees are for.
Viral family trees
These phylogenetic trees are essentially family trees for viruses. By mapping out the evolutionary relationships between different viral strains, it’s possible to find their common ancestors. Such trees also allow scientists to infer the direction of a viral host jump.
“If we find well-supported clades of strains all from host species A embedded within a host species B clade, we can be pretty sure that the virus jumped from host B to A,” said Balloux.
This was no simple task. The amount of data was huge. To make things even more challenging, the ecological diversity of viruses was much richer than previously classified using conventional methods. To overcome this problem, the researchers built their own phylogenetic tree that included 32 viral families based on genetic relatedness alone.
There was a problem with the metadata that the researchers had. Many viral genomes submitted to libraries lacked important information such as when, where, and which host the virus was taken from.
“In fact, 45% of sequences did not have host information and 37% did not have the date of sample collection. Additionally, the focus was mainly on viruses that infect humans, with 93% of all viral sequences associated with humans. This bias in sampling poses a big challenge to existing analytical approaches, and we spent a lot of time making sure that our results were not affected by it,” Cedric Tan, a Ph.D. student at UCL’s Genetics Institute and Francis Crick Institute, and lead author of the study, explained ZME Science.
Humans as major viral contributors
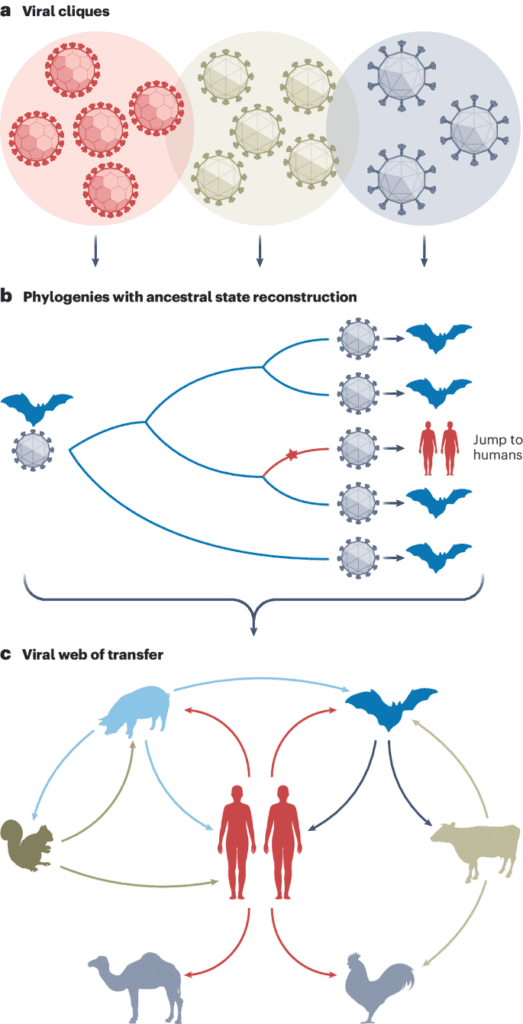
The researchers carefully reconstructed the evolutionary histories of viruses and studied genetic mutations acquired when they jumped between hosts. They found that human-to-animal transmission events are about twice as common as the reverse. Host jumps between animals that did not involve humans were even more common.
This once again emphasizes that humans are always part of nature — even though we may think otherwise. Instead, we are important parts of a complicated ecosystem, constantly exchanging pathogens with other species.
The study also looked into the genetic basis of these host jumps. They discovered that viruses often undergo significant mutations as they adapt to new hosts. Interestingly, viruses with a wide range of animal hosts showed fewer signs of such adaptive mutations. It seems that there is a natural flexibility in infecting various species.
“Another interesting finding is that viruses with a wide host range need fewer evolutionary changes to adapt to a new host. This likely explains the well-known pattern that the viruses of most concern for epidemics and pandemics tend to be generalists that can infect multiple hosts,” said Balloux.
Not well-studied but important to consider
The findings have wide-ranging implications, not only for understanding viral evolution but also for public health, conservation, and food security.
Viruses jumping from humans to animals could pose significant dangers to wildlife conservation, potentially endangering species and disrupting ecosystems. It’s currently not clear how much of a threat viruses jumping from humans are to other animals, as this relationship hasn't been studied much.
“The most well-studied example is human influenza A H1N1 (part of the seasonal flu) which regularly causes epidemics in livestock and sometimes wildlife. Other known cases of reverse zoonoses have been described for a series of human viruses, including hepatitis E, rotavirus, herpesviruses, and adenoviruses. One example is the outbreaks of human metapneumovirus and human respirovirus 3 in wild chimpanzees (endangered species) in Uganda, 2016-2017, causing several deaths in the Chimpanzee community. This is probably only a small part of the problem as disease outbreaks in wild animals are very rarely identified,” said Balloux.
“One of the first things we need to do to better understand the impacts of these host jumps is to strengthen efforts to monitor the genes of viruses in domestic and wild animal species. This will help us determine the variety of viruses infecting animals, how often host jumps occur, and the danger of new or recurring infectious diseases in these animals,” Tan stated.
Zoonosis and disease risk
Reverse zoonosis could also increase the chances of outbreaks among people. Completely new viruses can emerge through changes or a swapping of genetic material among different viruses infecting a host at the same time. This swapping process, called reassortment, is especially risky.
A good example is the movement of viruses from pigs to humans, as pigs can mix different viruses together and then pass on new changed strains to humans. In 2009, the H1N1 flu virus killed between 151,700 and 575,400 people globally in its first year. However, this virus, which transferred from animals to humans, contained gene sections from four different sources: humans, birds, the North American pig, and the Eurasian pig.
It's not clear how much reverse zoonosis raises the risk of pandemics or major outbreaks in general. Most newly appearing zoonotic diseases have come from wildlife, not livestock or pets. However, the connections between humans and other species are extremely complex, which means we need much more gene surveillance than we currently do.
The results were featured in the journal Nature Ecology & Evolution.
Was this helpful?
Related Posts
- Scientists turn to the troubled streets of Gotham to understand community resilience
- Different types of foods are associated with different types of stroke
- Basic income in another Dutch town: each person receives up to $1,450/m unconditionally
- Pot twist: Cannabis component helps fight addiction in new study









