Since the advent of modern DNA sequencing technology, biological research and discoveries has been dramatically accelerated. It’s absolutely instrumental to genetic research nowadays, which among other great achievements, has lead to the sequencing of the human genome. The methods and technologies involved in DNA sequencing are terribly complex, however, and usually require sophisticated research laboratories. What if you could simplify the process?

Oxford Nanopore (ON) had this idea in mind for some time, and recently unveiled an extraordinary product the company has completed developing – a fast, portable, and disposable nucleotide sequencer the gets powered via USB and runs analysis on the same computer it gets plugged in. Extreme costs are promised to be alleviated once this products gets introduced on the market, eliminating the need for highly expensive facility usage for small projects and offering the possibility to perform genetic analysis on the go when needed.
The MinION, as it’s been dubbed by ON researchers, doesn’t need polymerase chain reaction (PCR) or other DNA amplification technique for optimum sensitivity, and can sequence up to 150 million base pairs within its six hour working life. The device accepts samples of blood, plasma, and serum for an immediate analysis.
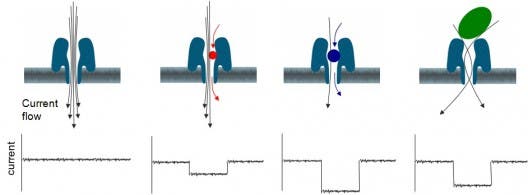
MinION’s centerpiece is its nanopore port. A nanopore is basically an organic molecule with a very narrow hole, just a few nanometers in width. This nanopore is embedded inside two molecule thick synthetic polymer membrane, which has a very high electrical resistance, such that the nanopore hole forms a path from one side of the membrane to the other. Through the nanopore hole electrophysiological fluid is inserted, which has its volume divided in half as a result of a specific geometry.
When passing through the variable geometry in the nanopore hole, the electrophysiological fluid is swept by an ionic current which causes a voltage difference. Each molecule, including DNA or RNA, has its own characteristic voltage, and thus using this technique the MinION can detect and identify the sample. Of course, the MinION’s main purpose is that of sequencing DNA, so the device is optimized to differentiate the four nucleobases (adenine, cytosine, guanine, and thymine) which encode genes in DNA. To analyze the DNA, the MiniON uses strand sequencing.
The device’s sensing electronics has 512 nanopores embedded onto its sample chip, resulting in a total strand reading rate of about 7500 bases/second. During its limited 6 hours operation life time, the MinION can read 150 million bases; enough to read small chromosomes or bacteria genome. Check out this excellent video from Oxford Nanopore explain in great detail how the MinION works and how the DNA sequencing process unfolds.
Nanopore DNA sequencing from Oxford Nanopore on Vimeo.
The researchers working on the device have already tested the device successfully, after sequencing the genome of the lambda bacteriophage – 48500 base pairs in length. Clive Brown, the Chief Technology Officer of Oxford Nanopores, has been cited during the product’s announcement at AGBT 2012, that the MinION might be introduced on the market with a $900 price tag!
“The exquisite science behind nanopore sensing has taken nearly two decades to reach this point; a truly disruptive single molecule analysis technique, designed alongside new electronics to be a universal sequencing system. GridION and MinION are poised to deliver a completely new range of benefits to researchers and clinicians,” said Dr Gordon Sanghera, CEO of Oxford Nanopore. “Oxford Nanopore is as much an electronics company as a biotechnology company, and the development of a high-throughput electronics platform has been essential for us to design and screen a large number of new candidate nanopores and enzymes. Our toolbox is customer-ready and we will continue to develop improved nanopore devices over many years, including ongoing work in solid state devices.”
press release / via Gizmag